User Guide
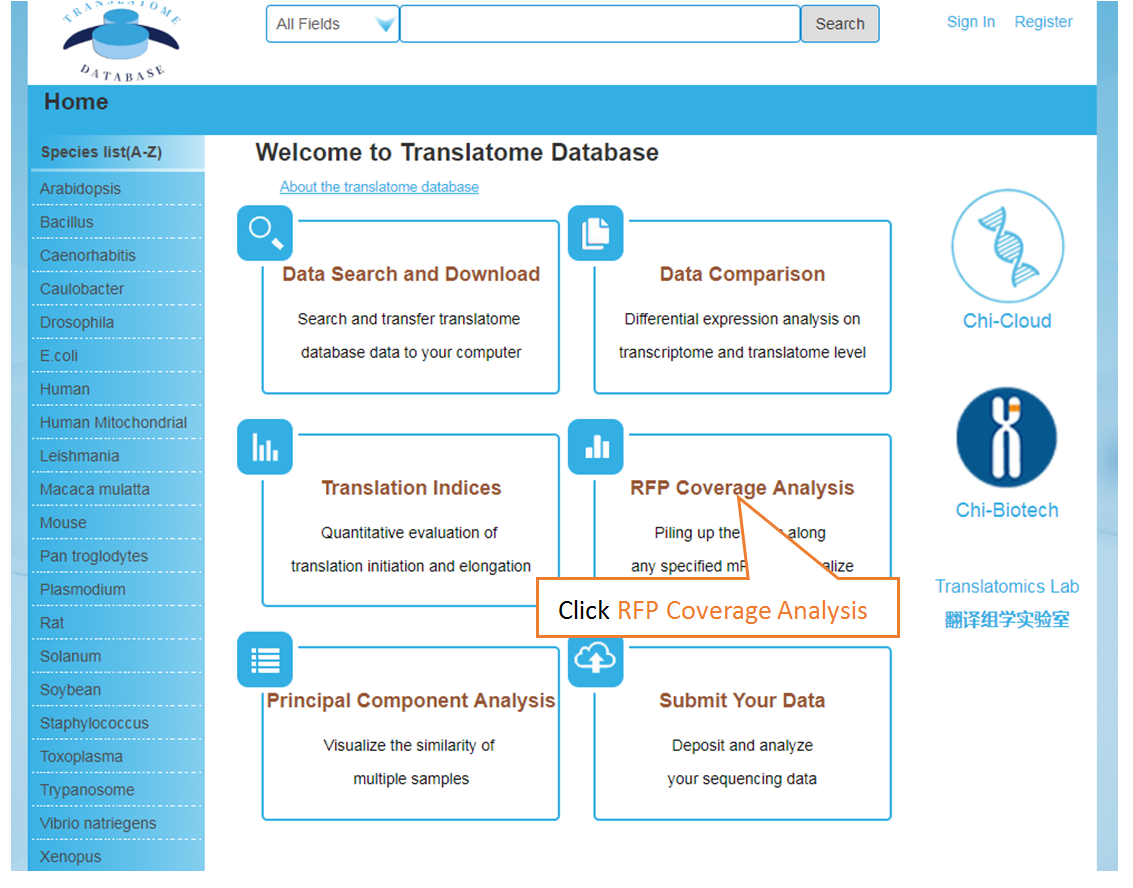
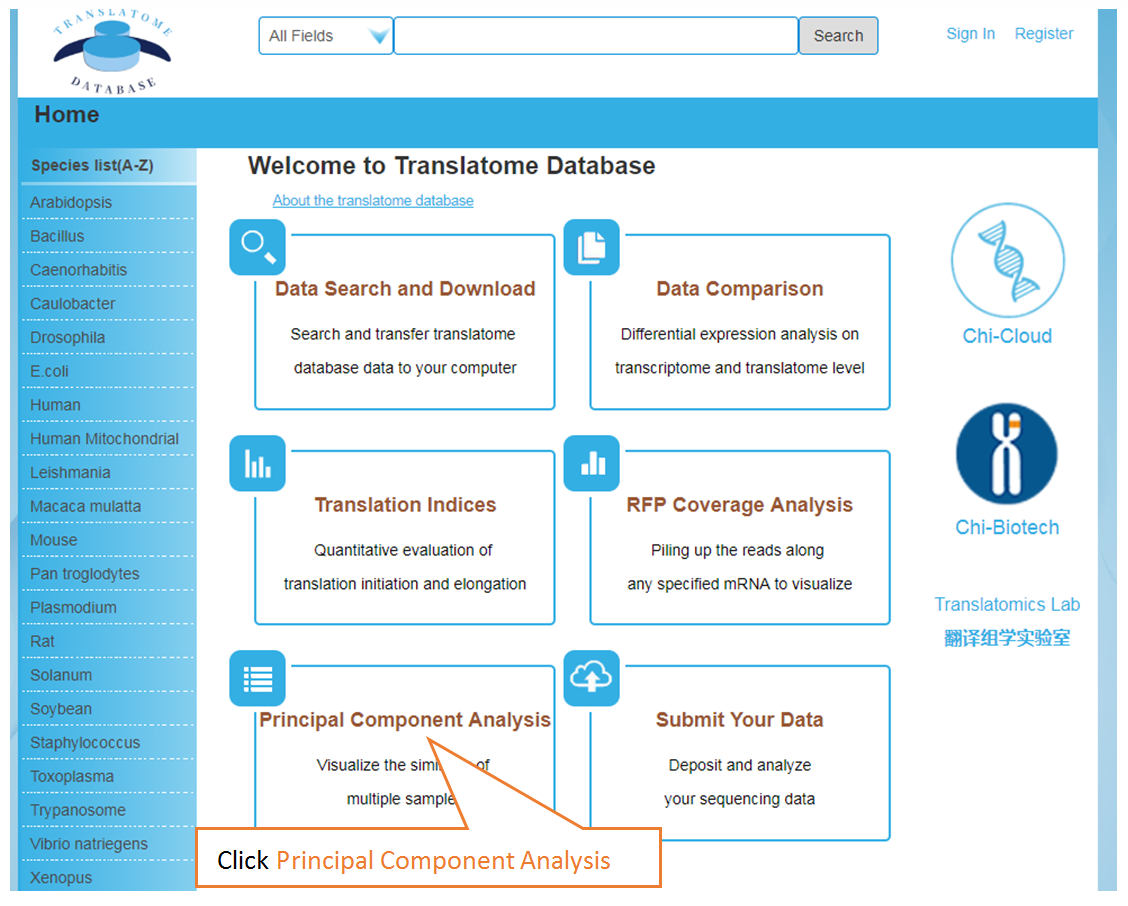
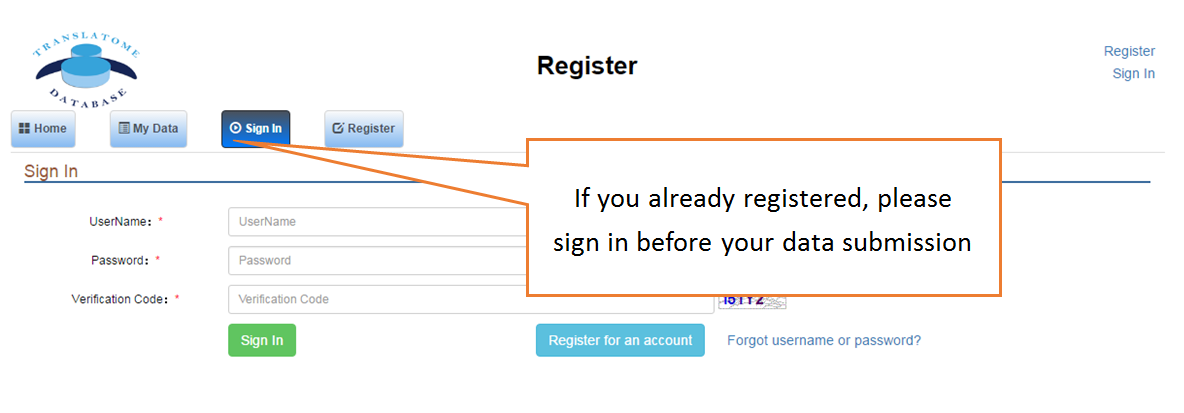
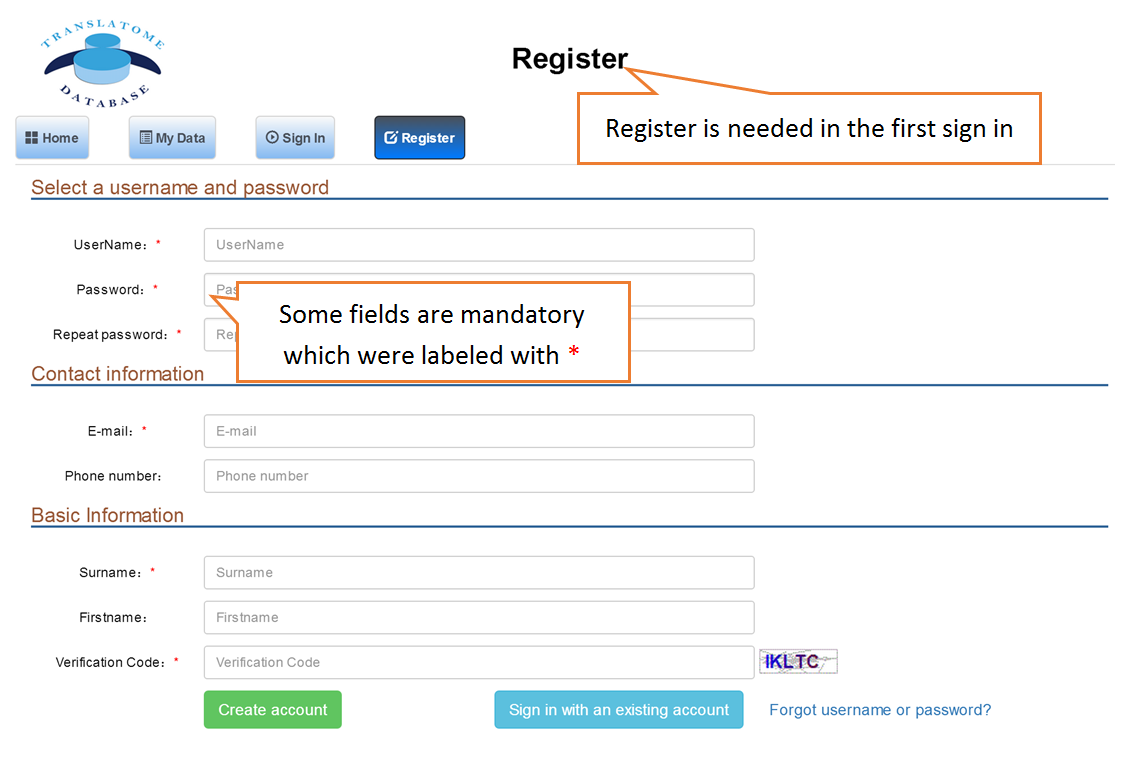
The Translatome Database collects the RNC-seq, Ribo-seq and the corresponding mRNA-seq data, and provides a unified analysis pipeline for integrated analysis. The current version includes 3,288 Ribo-seq, 40 RNC-seq and their 2,415 corresponding mRNA-seq datasets of 23 species which were uniformly analyzed by the hyper-accurate, experimental-verifiable and robust FANSe3 algorithm. The main sections of the website are Data Search and Download, Submit Your Data, Data Comparison , Translation Indices , RFP Coverage Analysis and Principal Component Analysis. Here is a quick–start guide to running the translatome database.
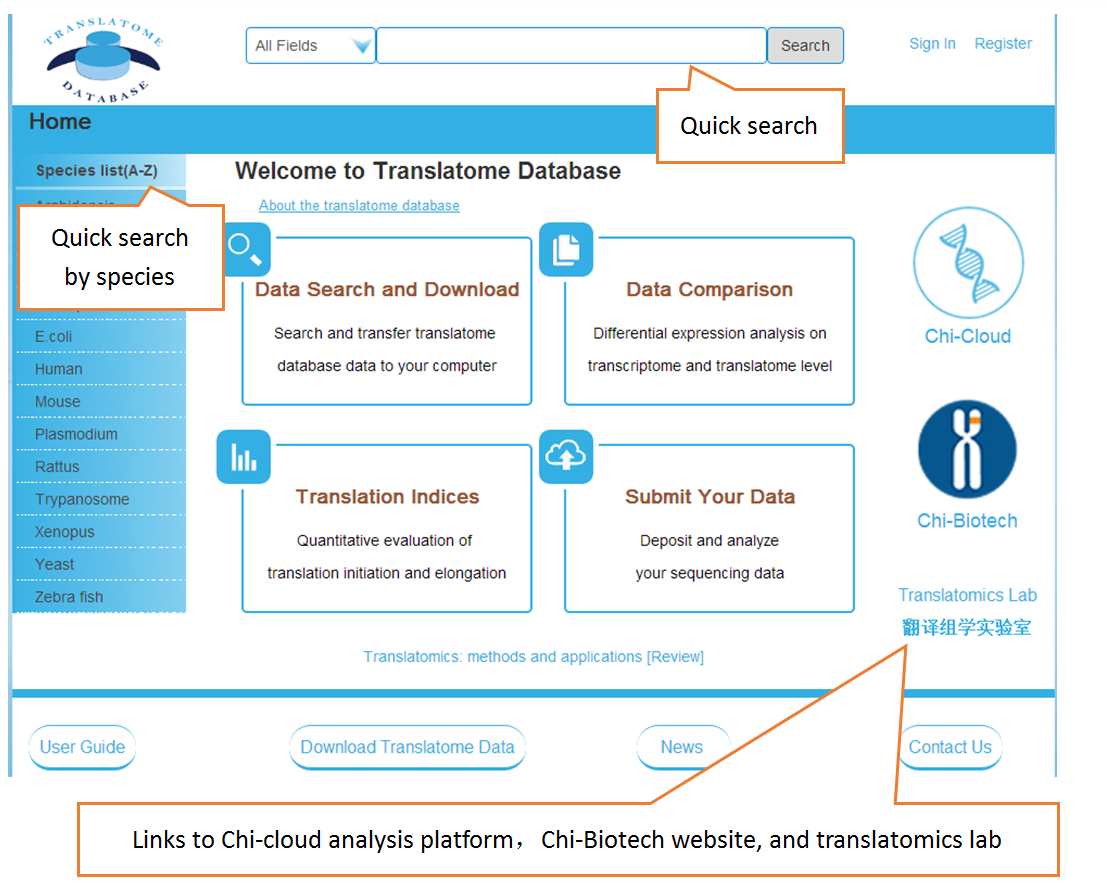
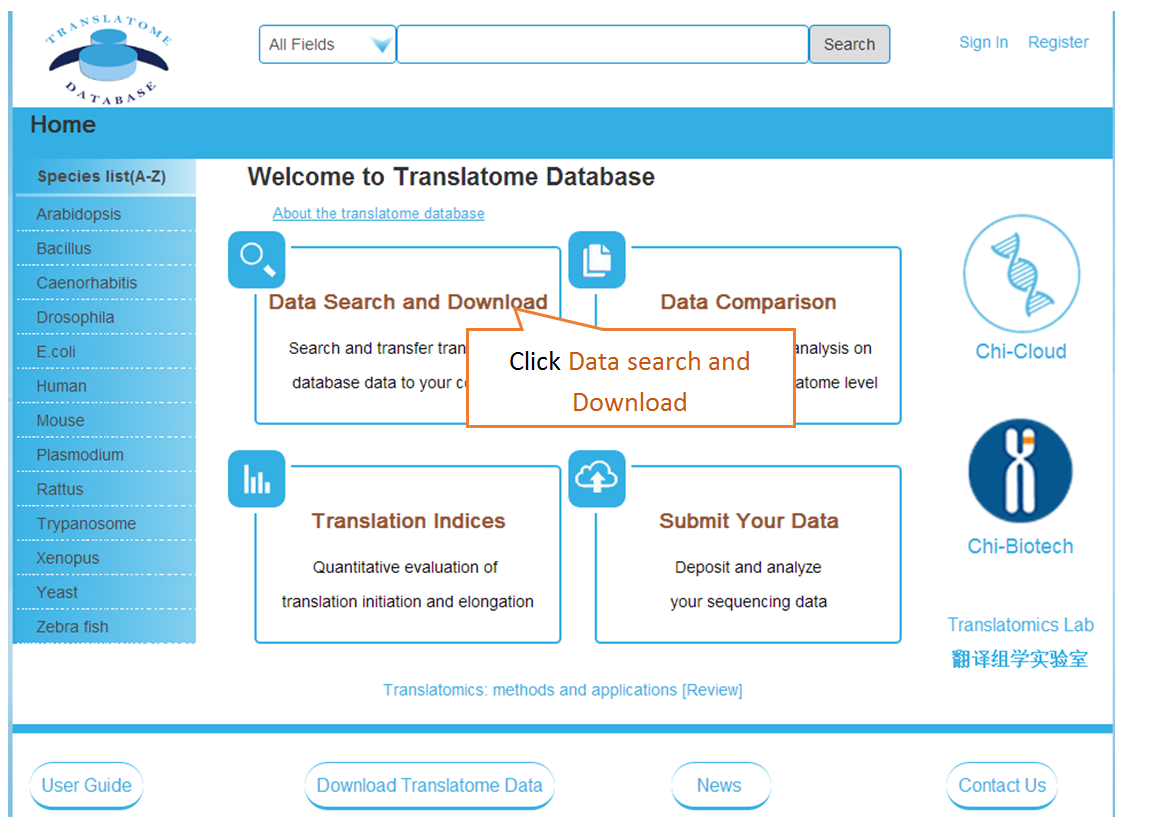
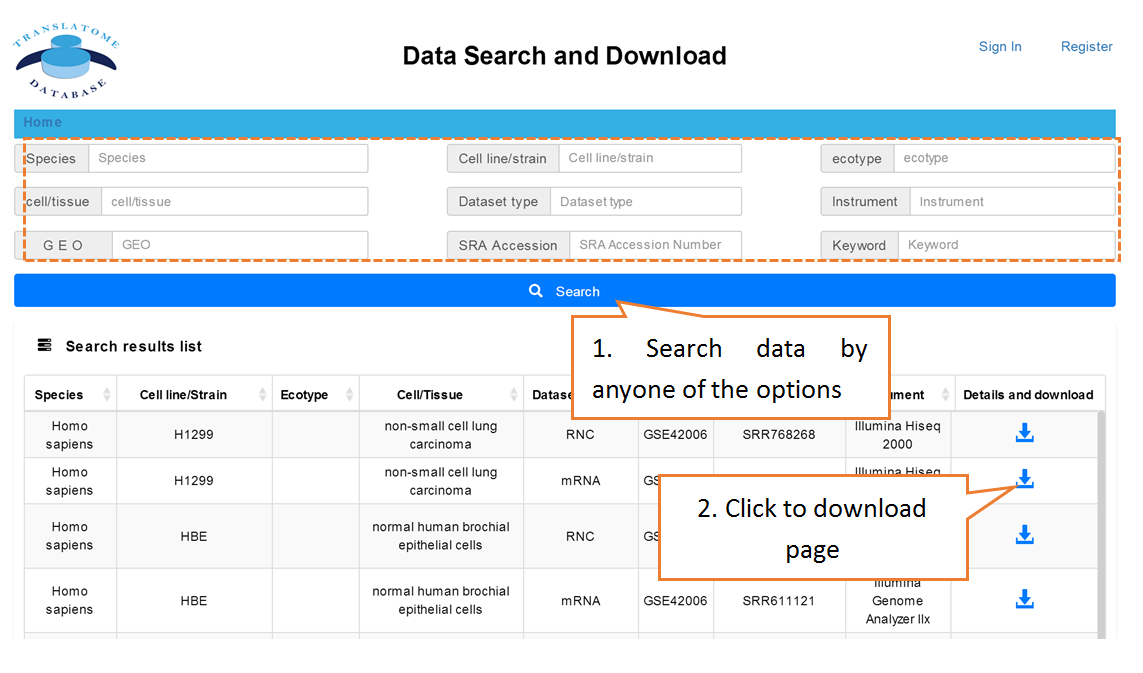
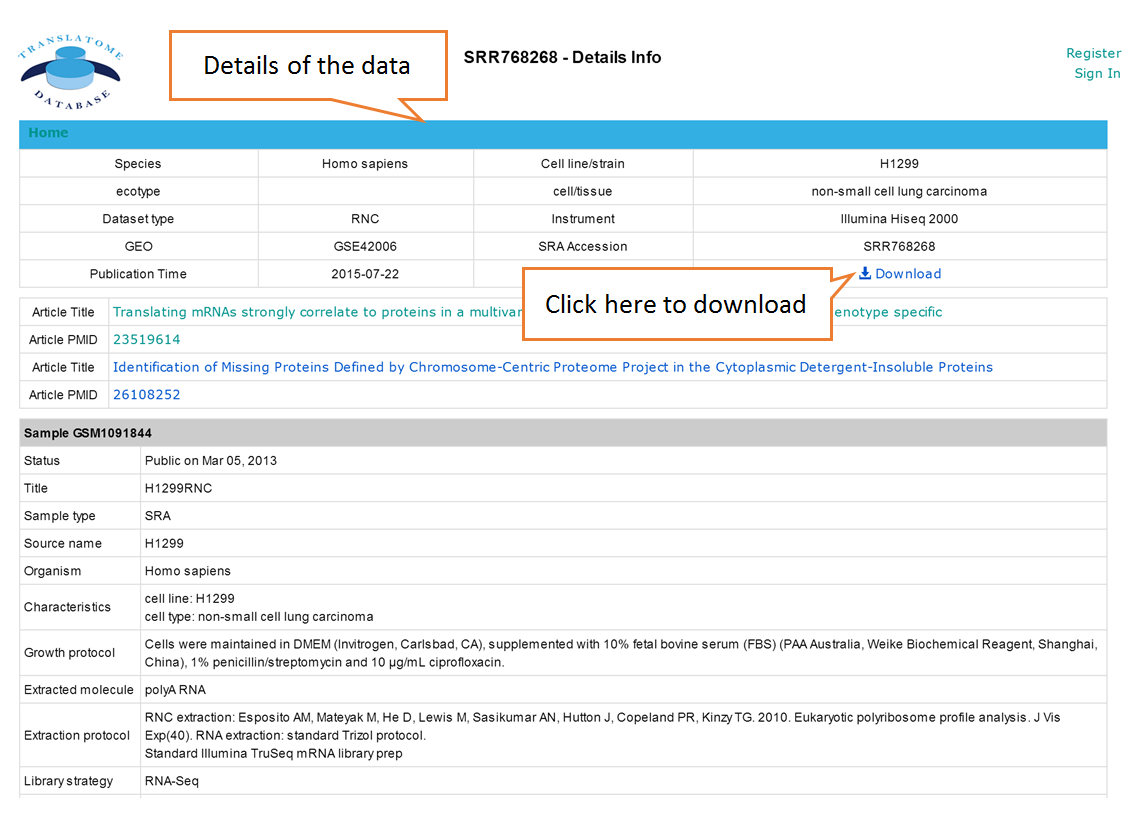
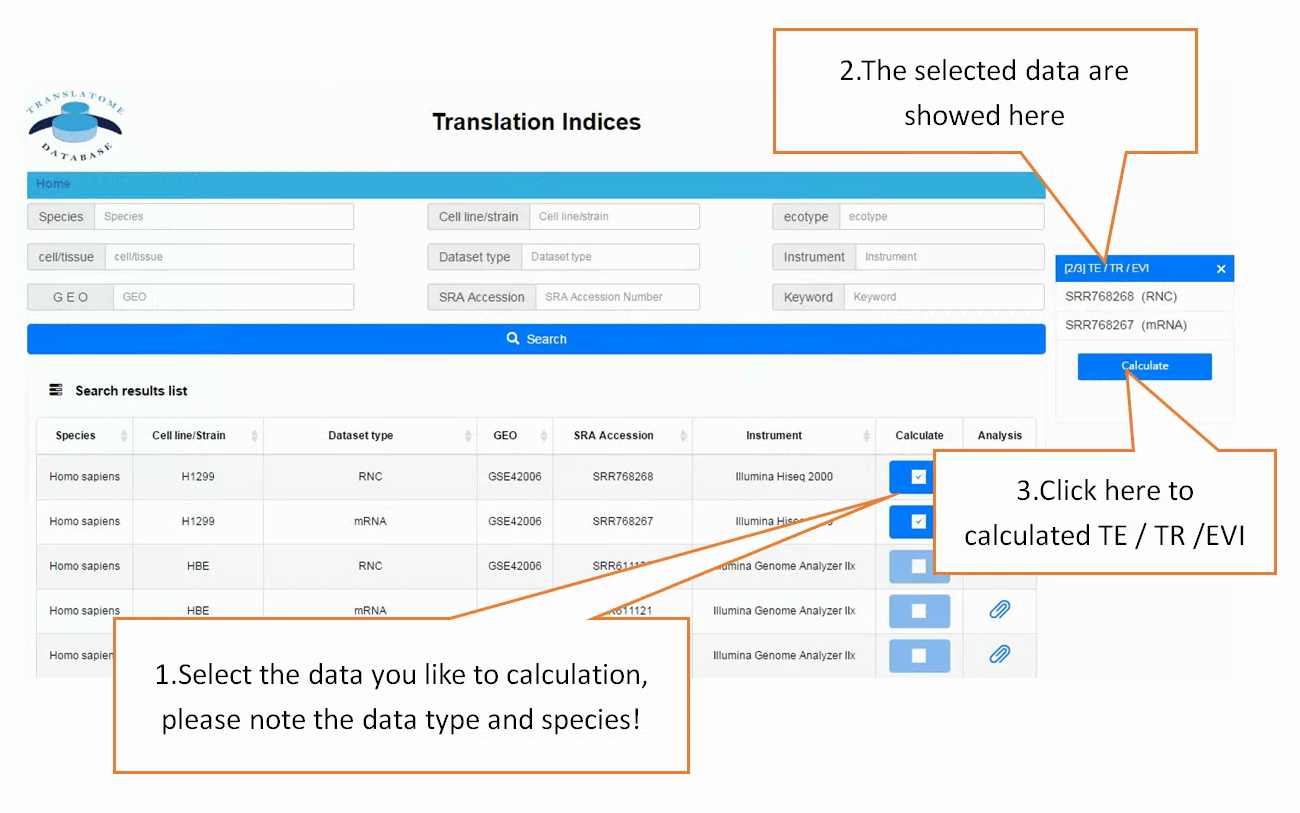
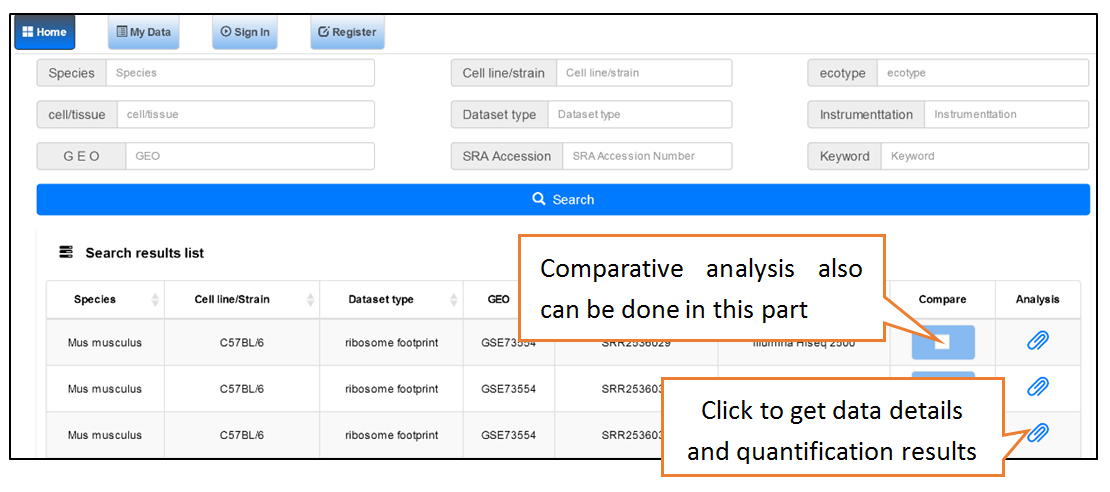
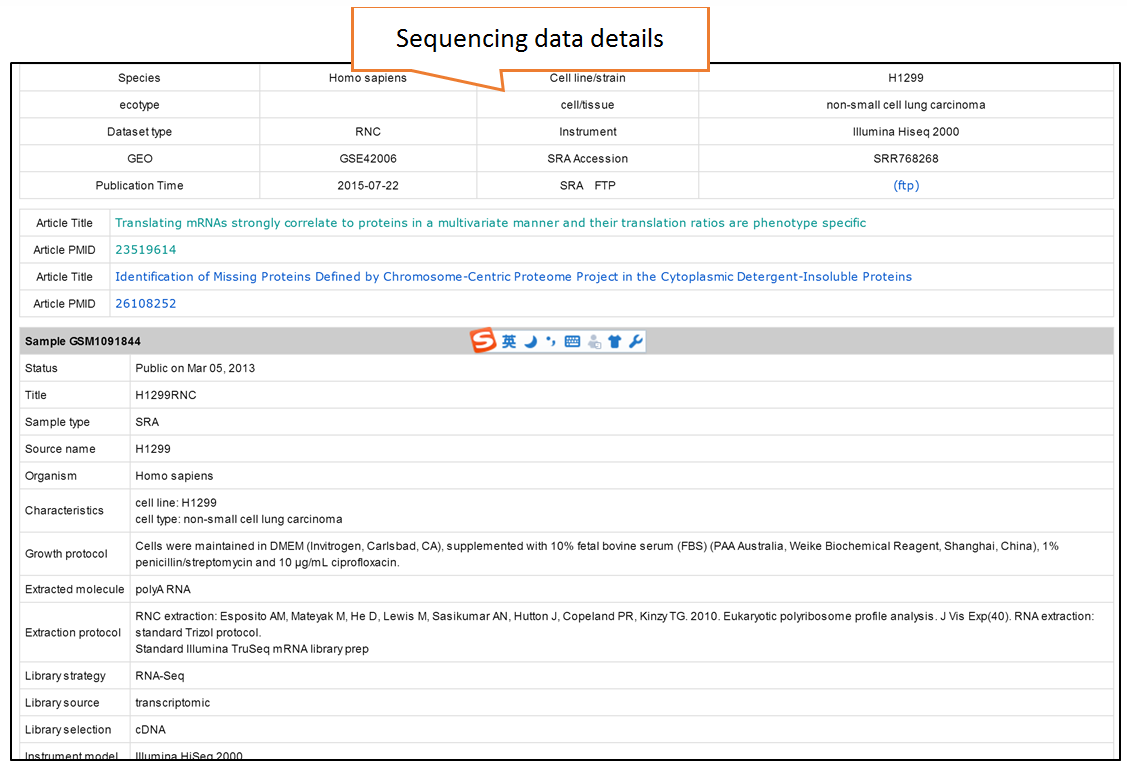
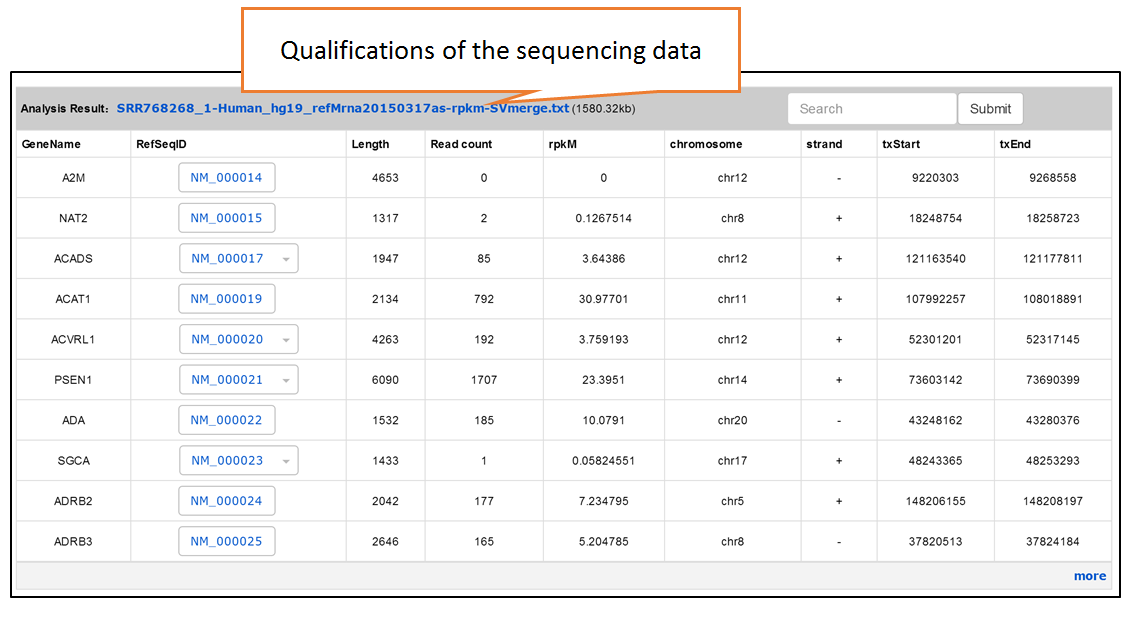
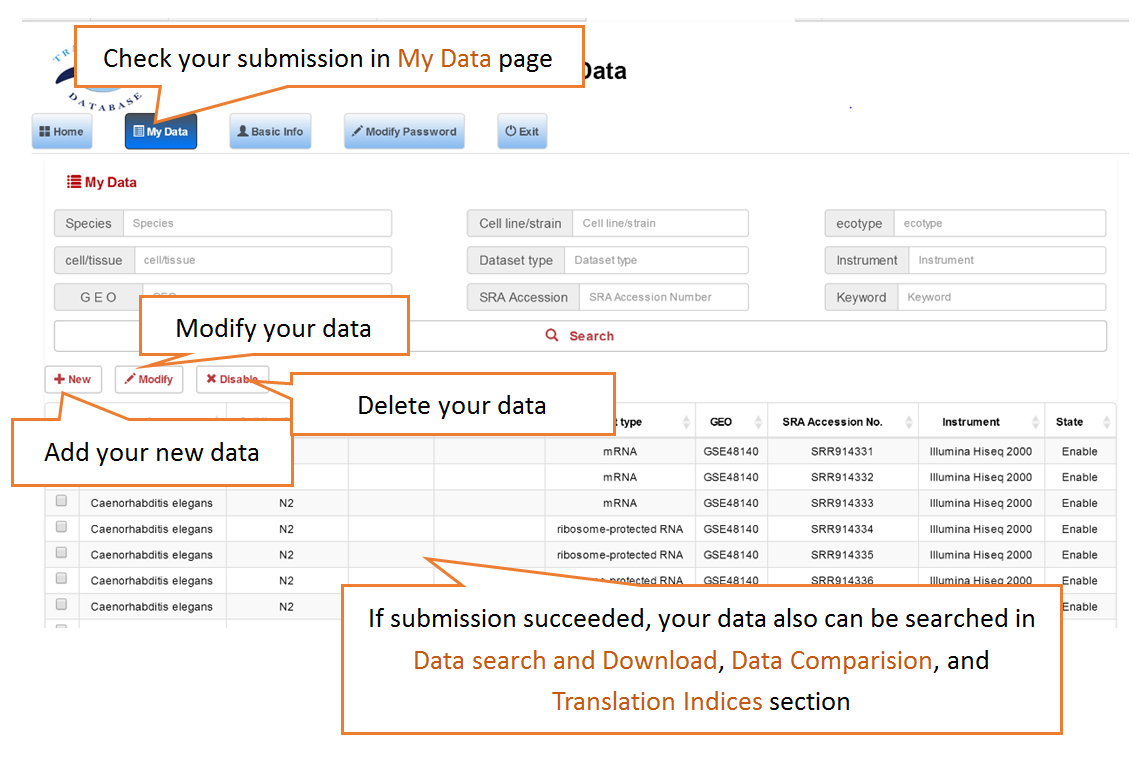
Users can search the sequencing datasets using 9 fields of the metadata, including species, dataset types, GEO numbers, sequencing instrument, cell line/tissue types, SRA accession number and so on. One or more key words are accepted to precisely get the desired datasets.
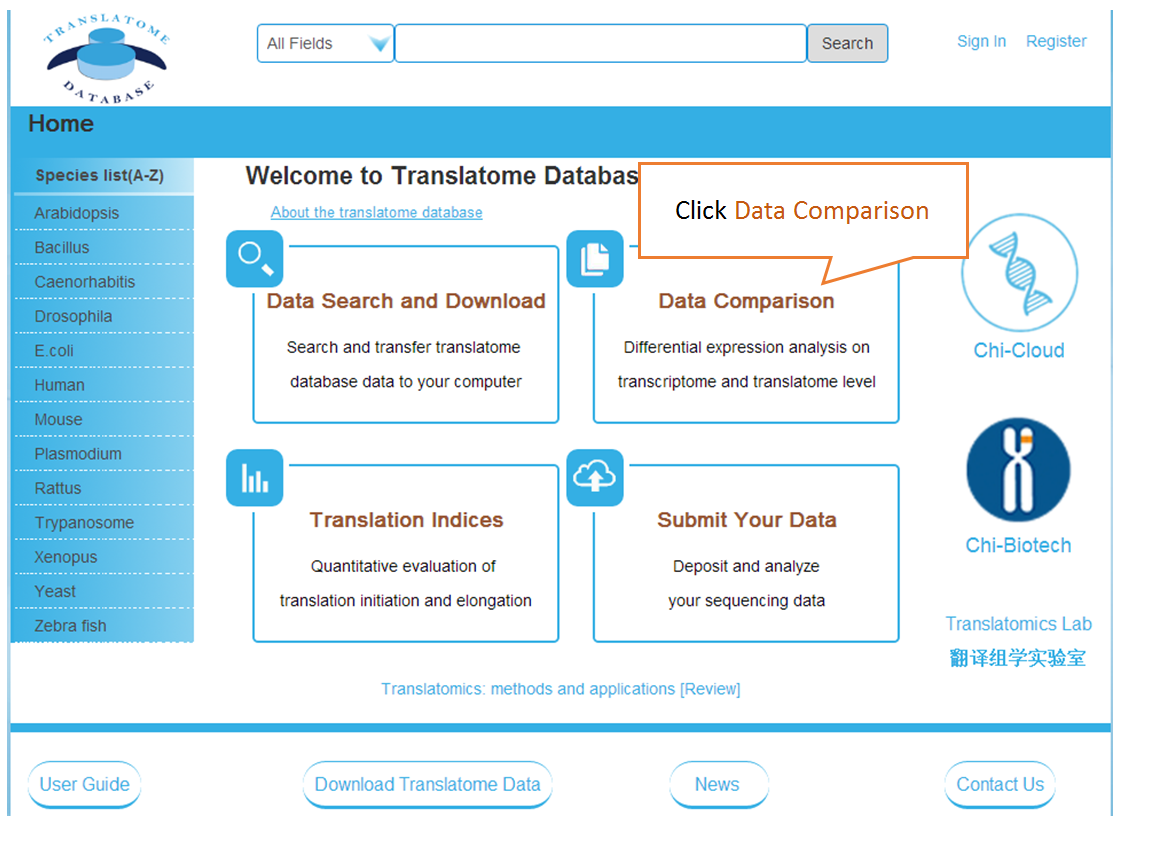
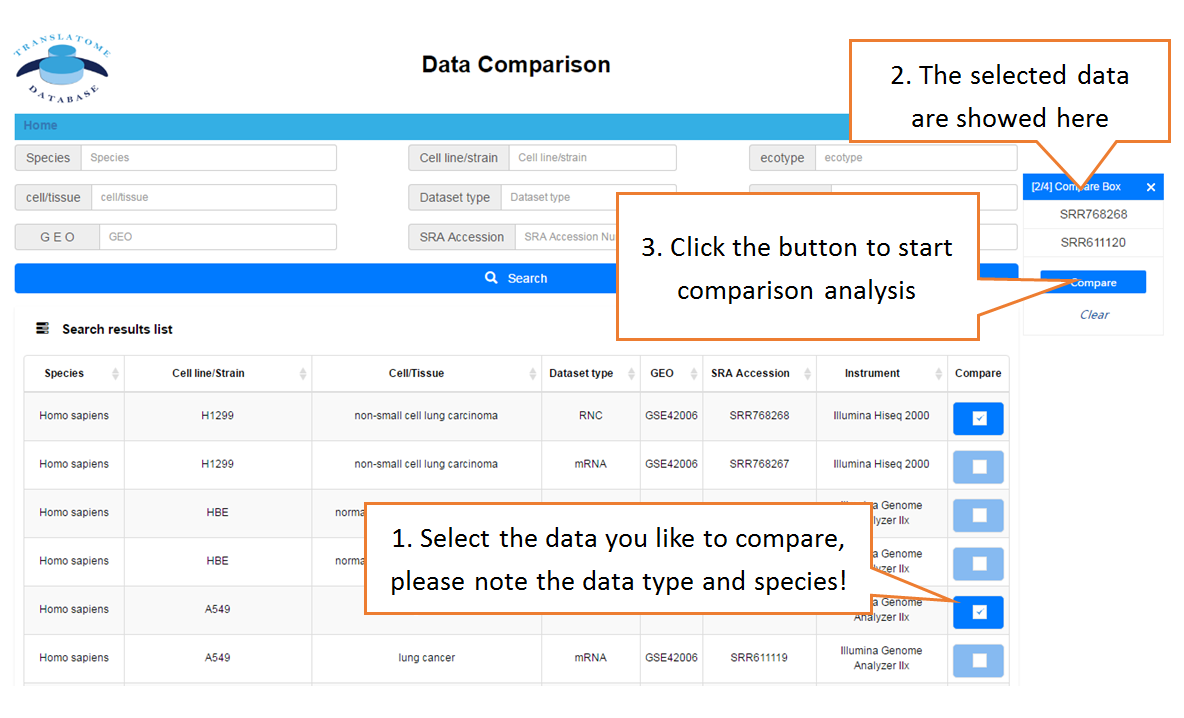
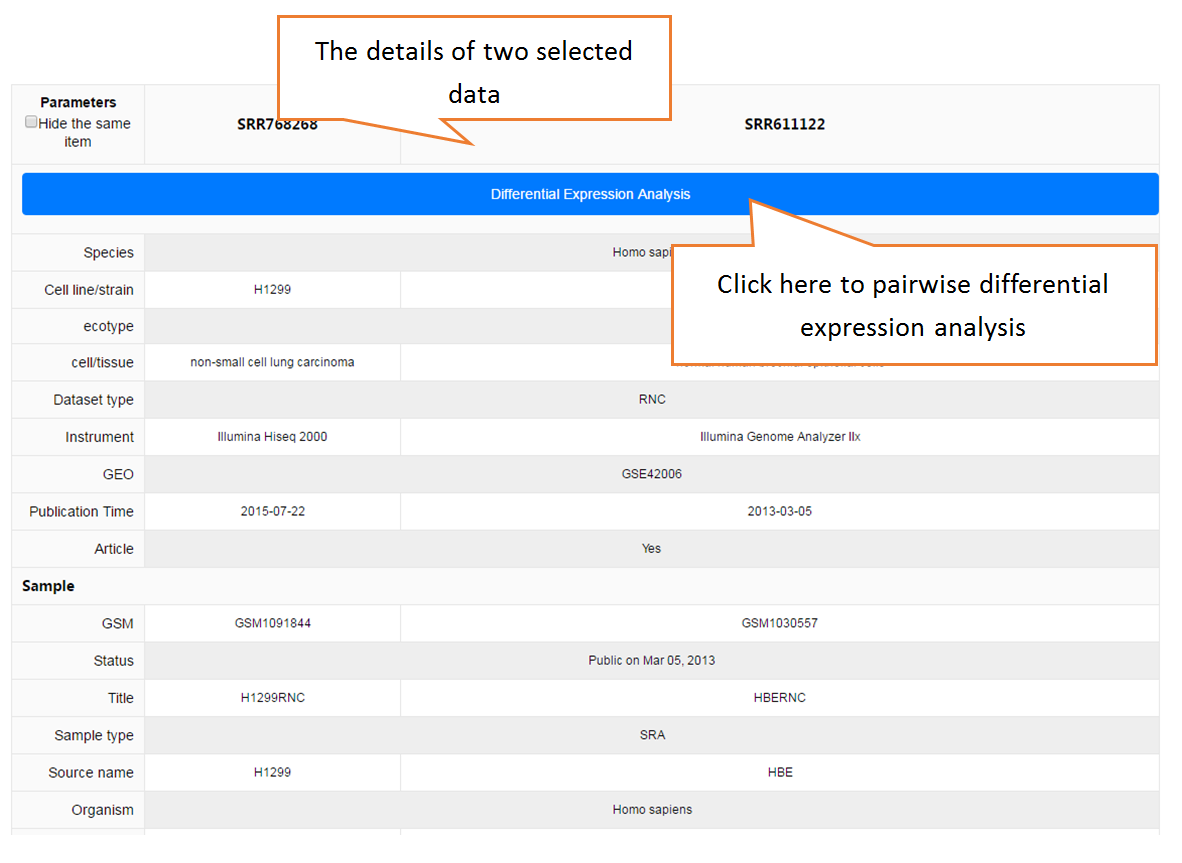
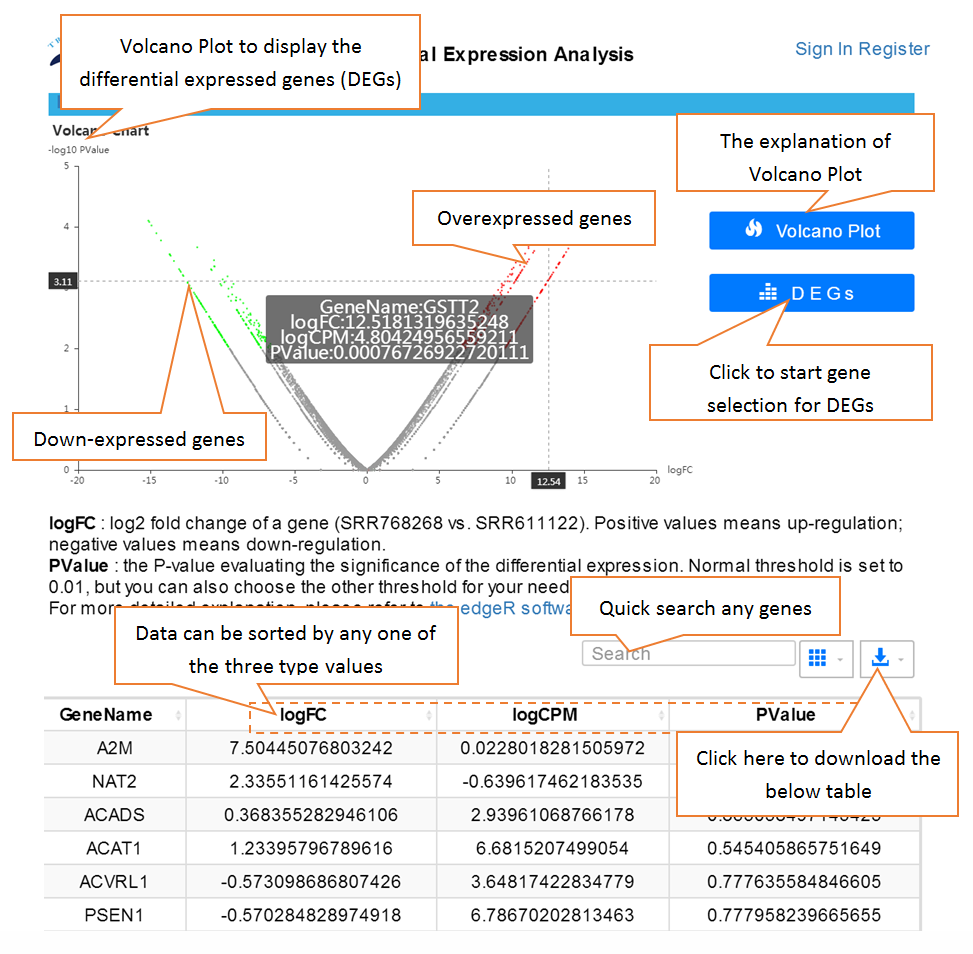
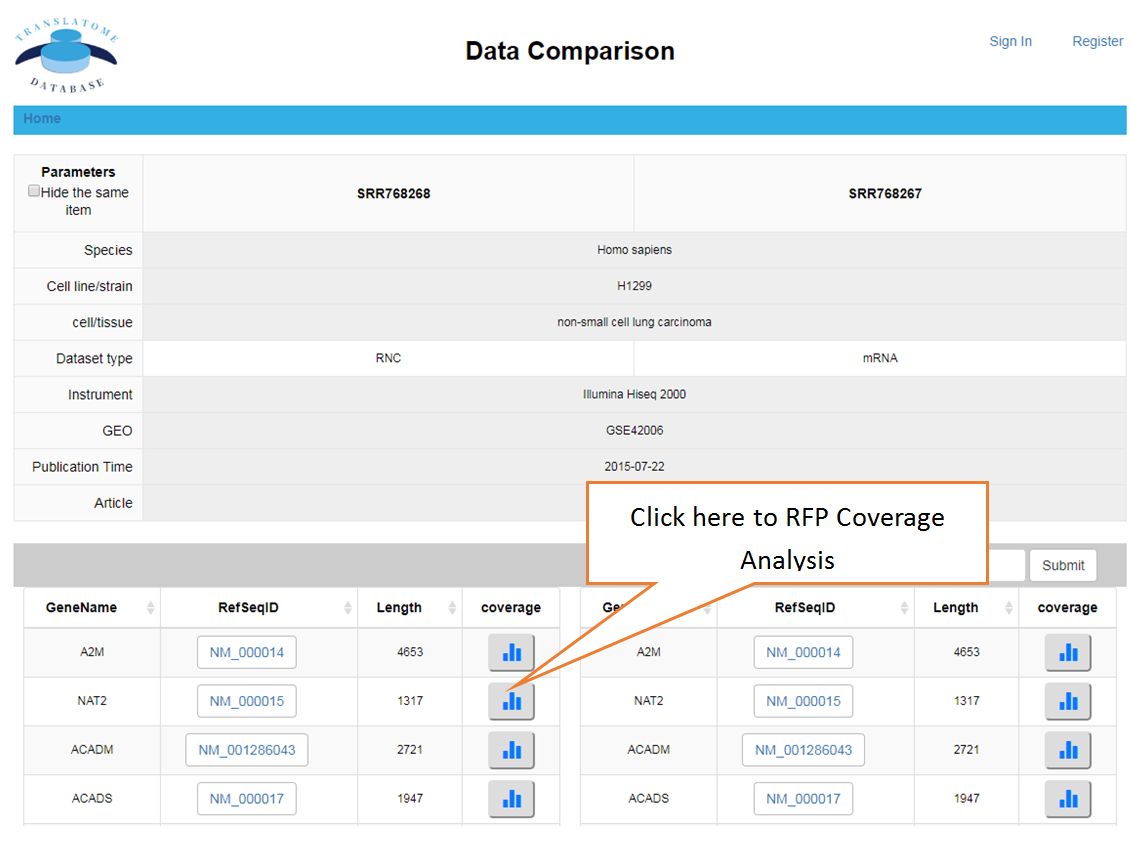
Data comparison section performs Differential gene expression (DGE) analysis between any two datasets of the same species and type, both on transcriptome and translatome levels.
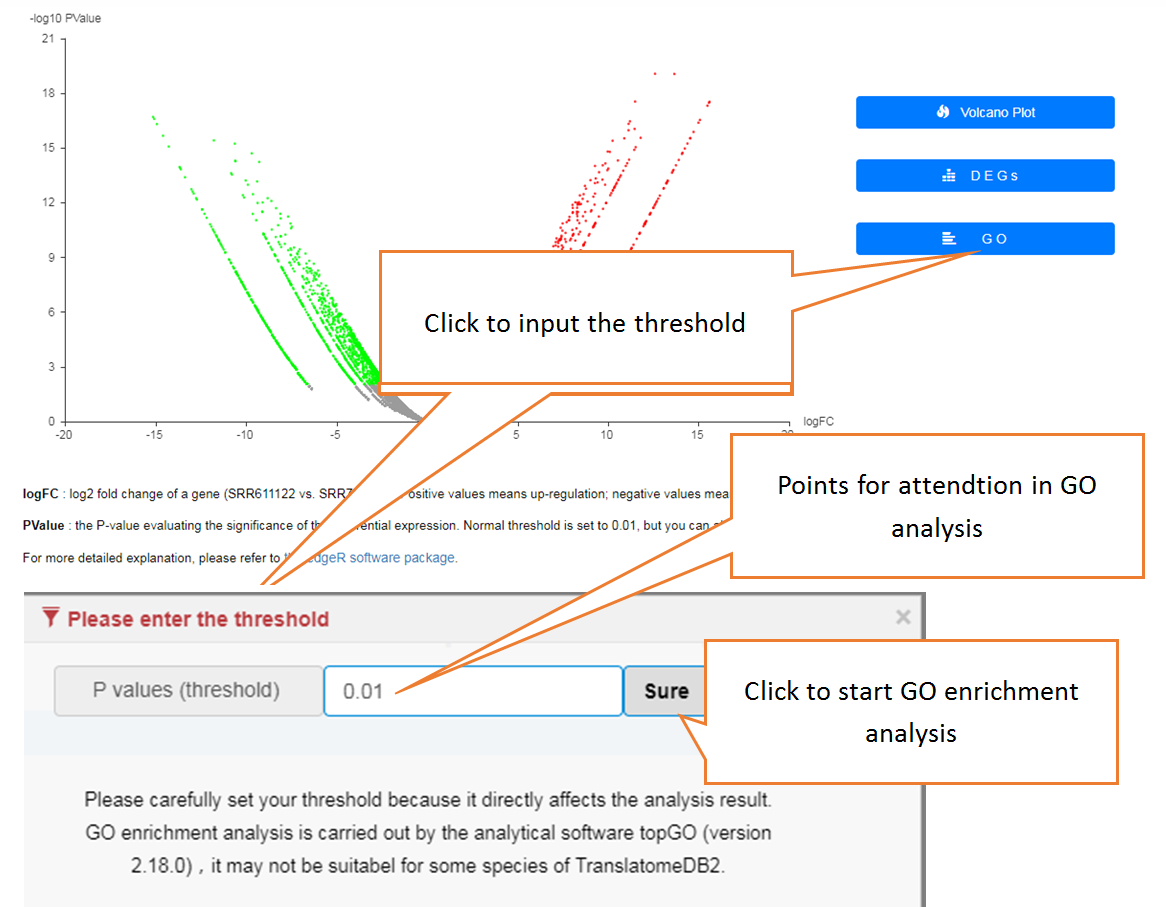
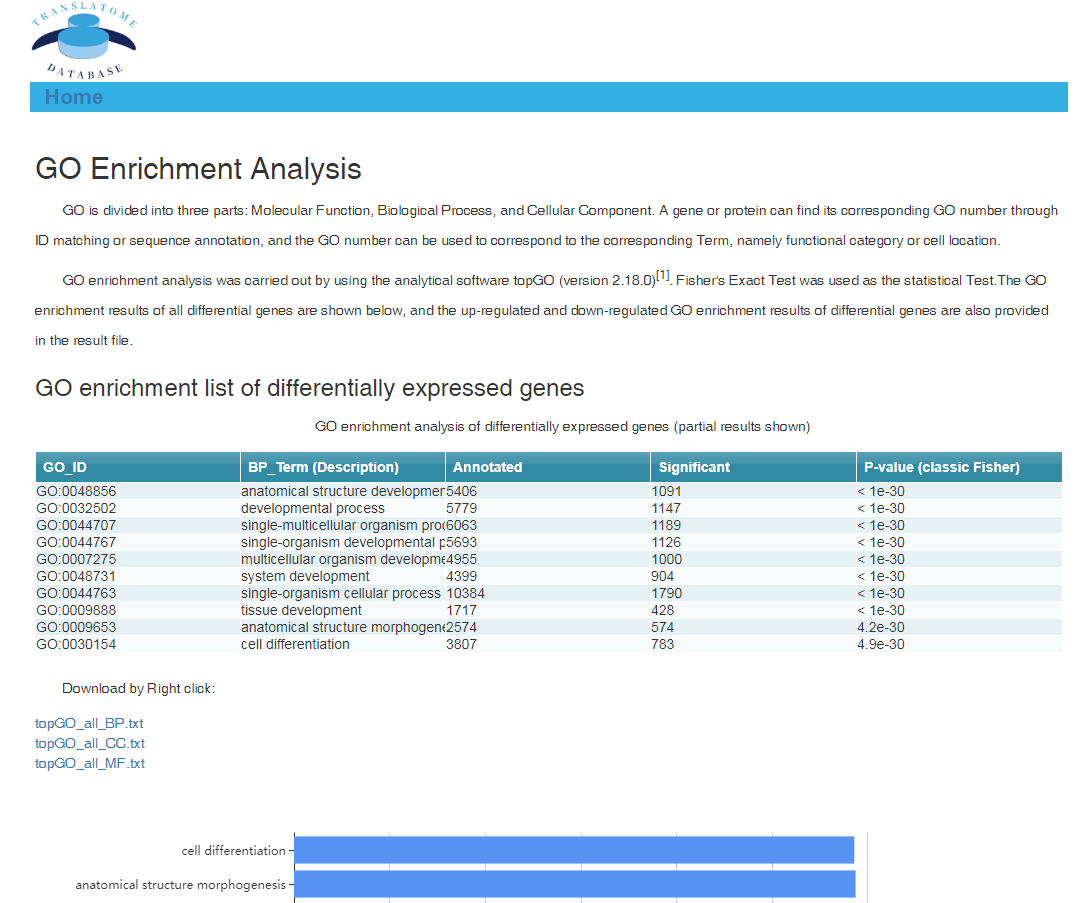
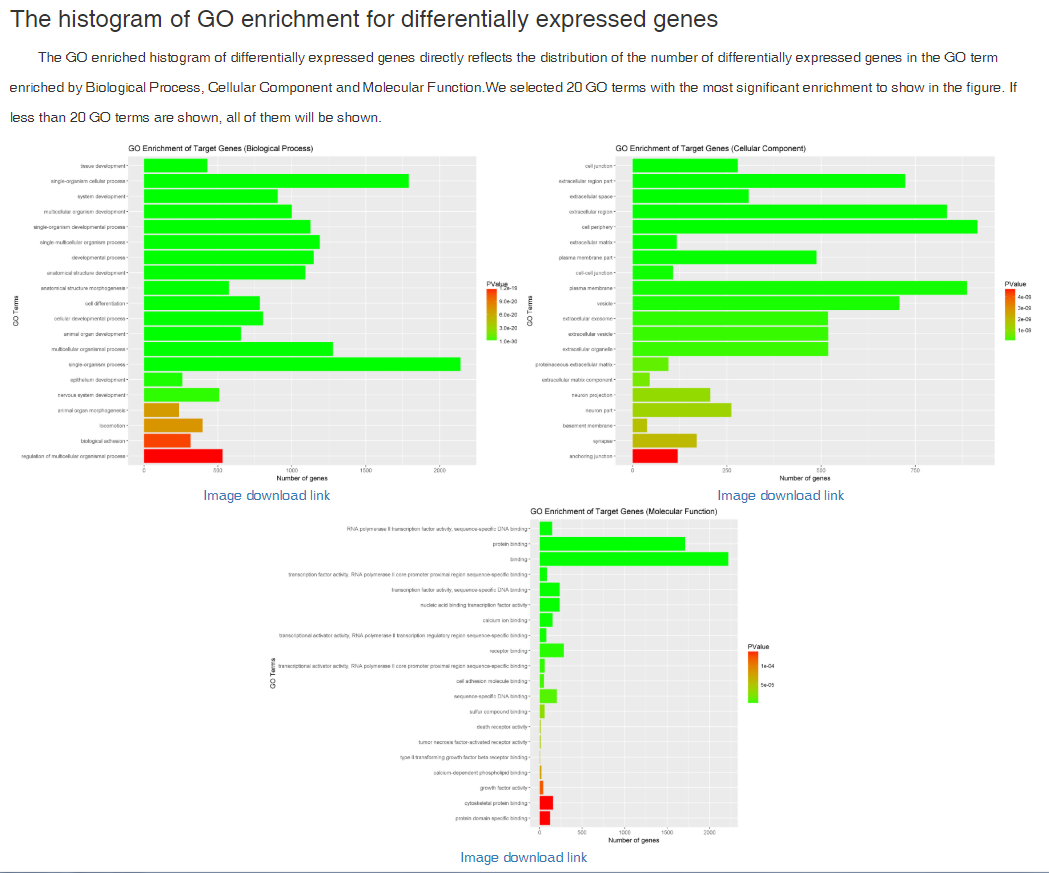
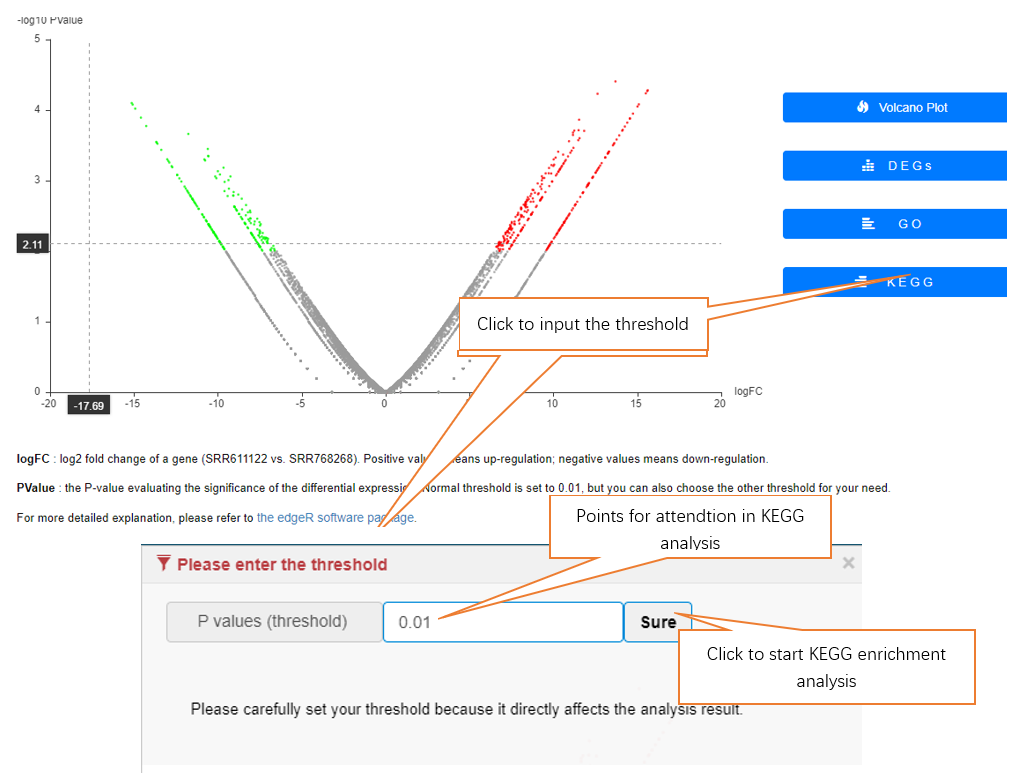
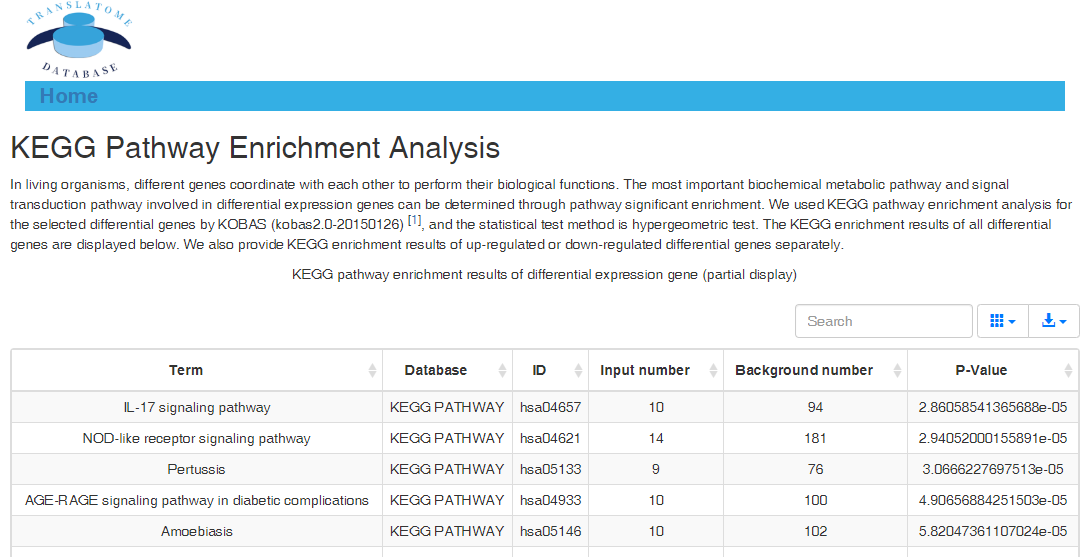
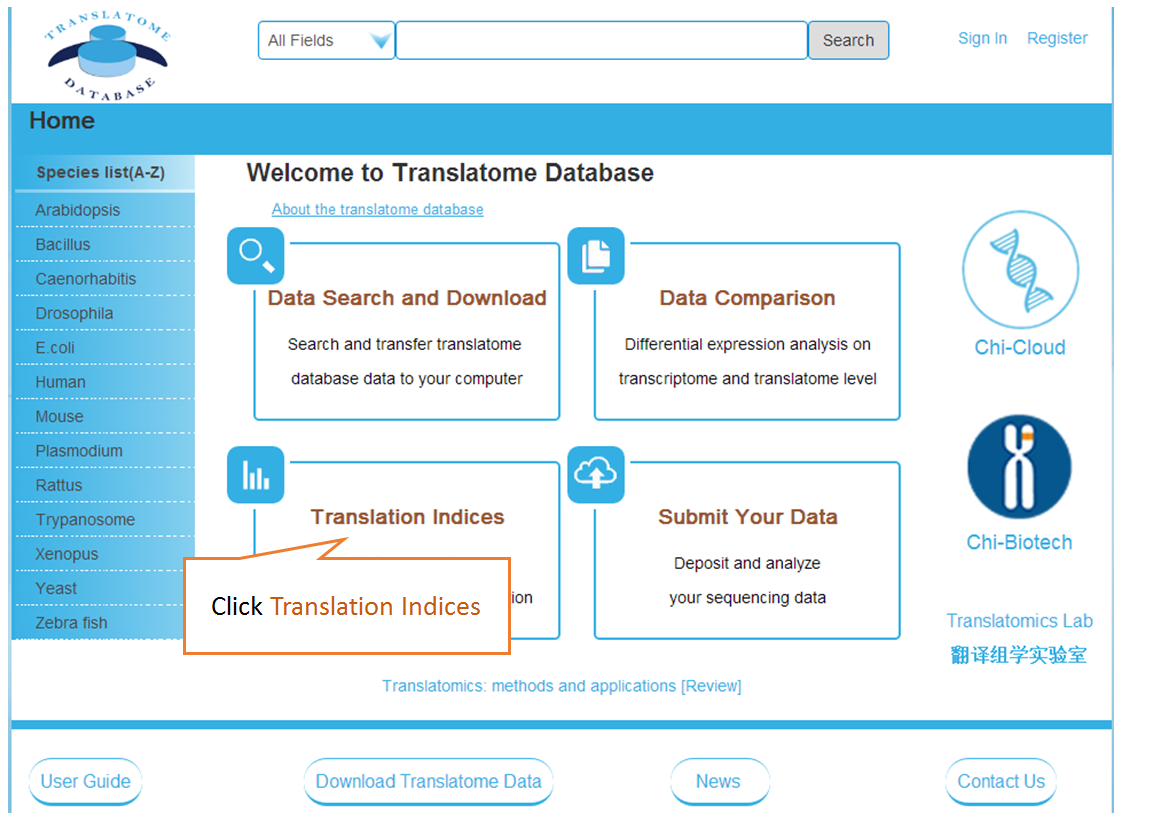
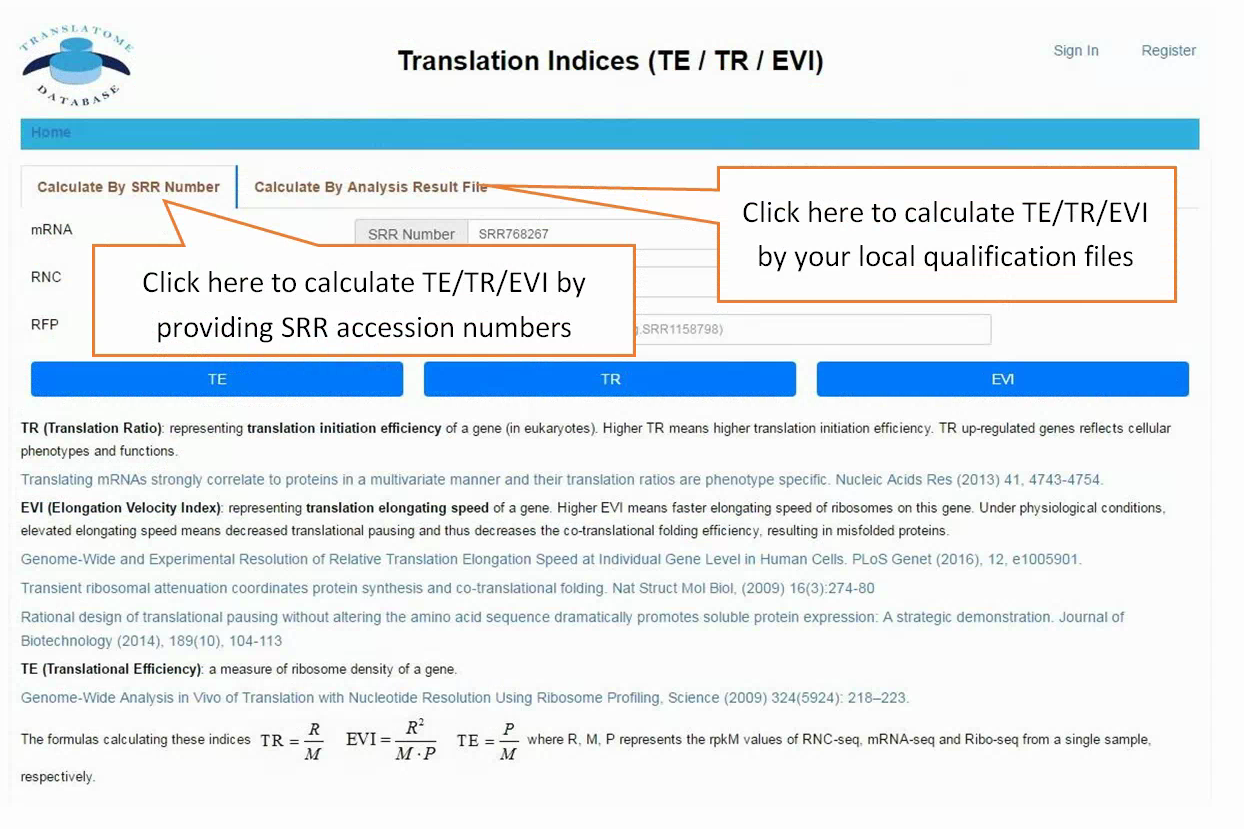
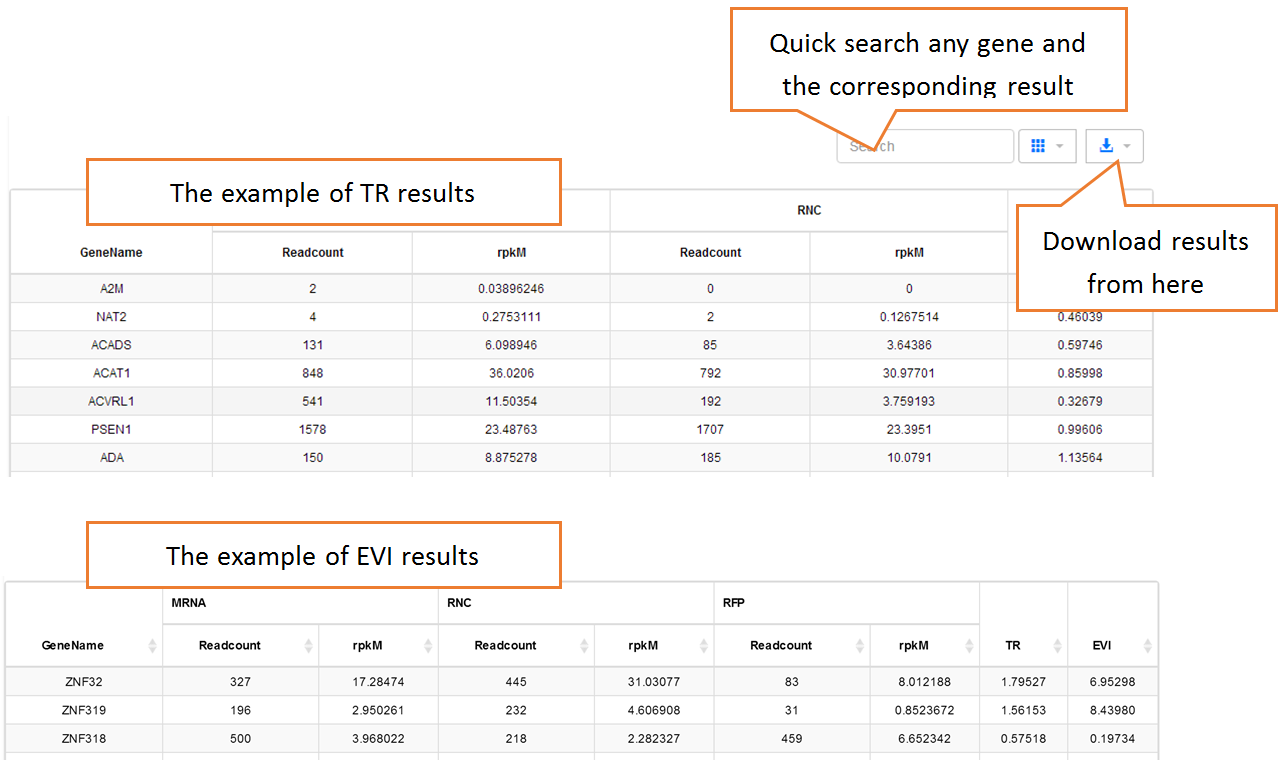
In translation indices section, the translation indices TR and EVI can be calculated for single sample to quantitatively evaluate translational initiation efficiency and elongation velocity, respectively.
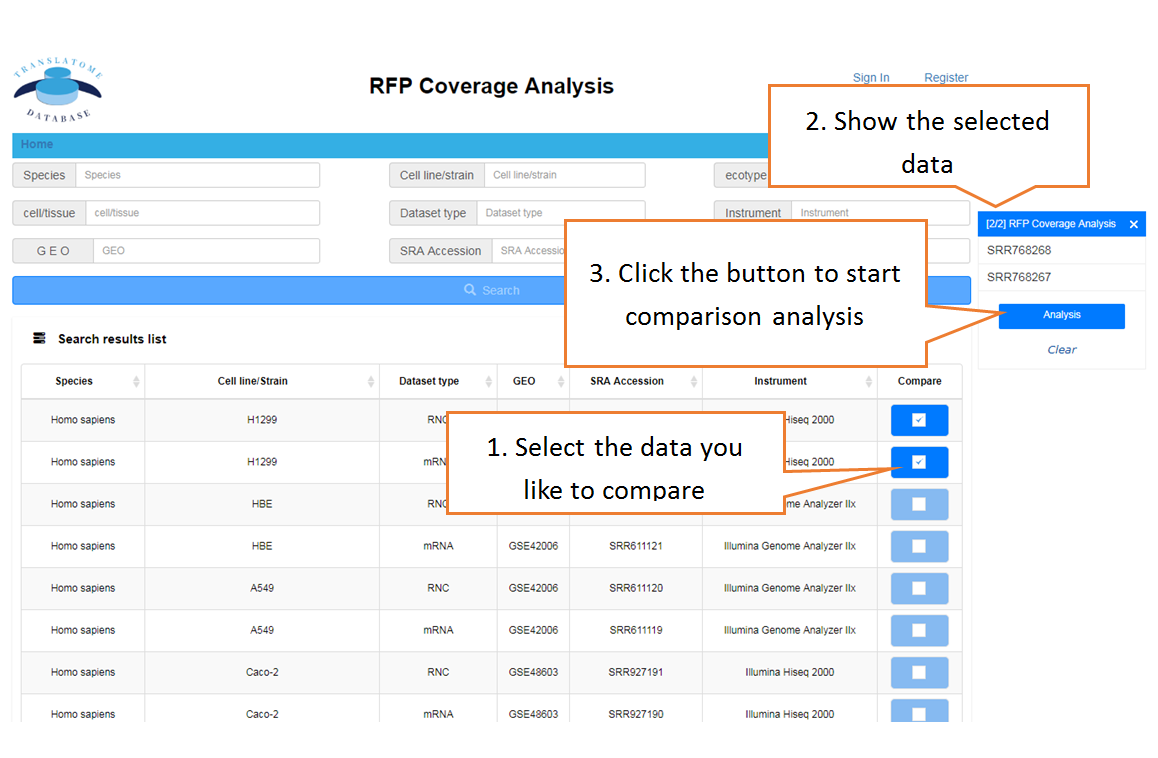
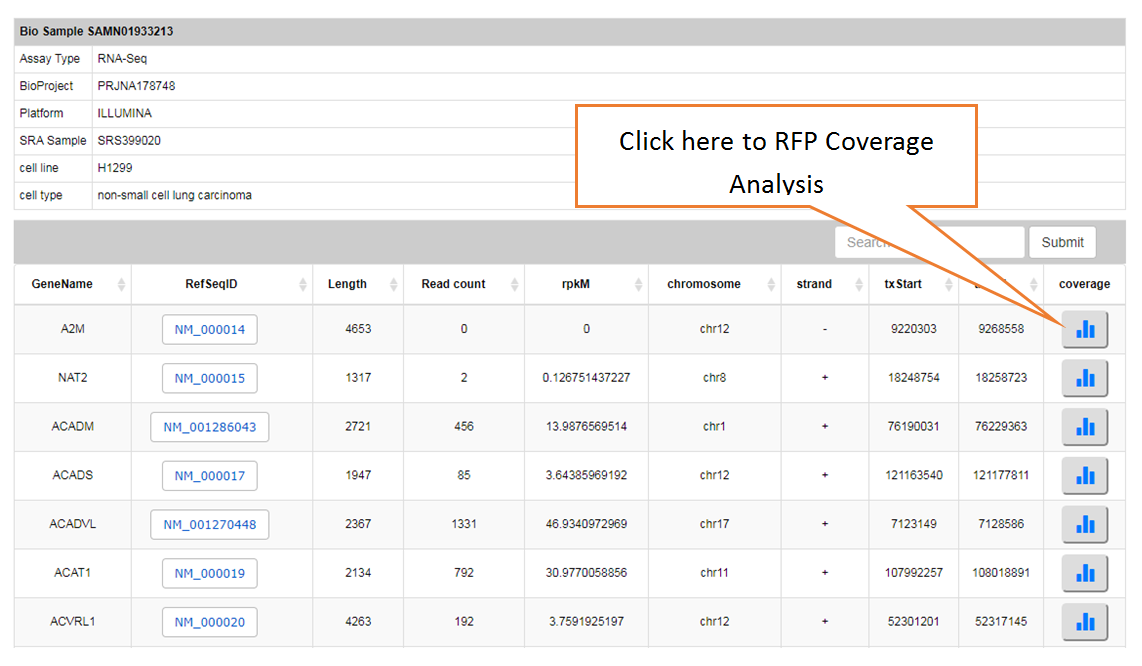
Piling up the reads along any specified mRNA to visualize
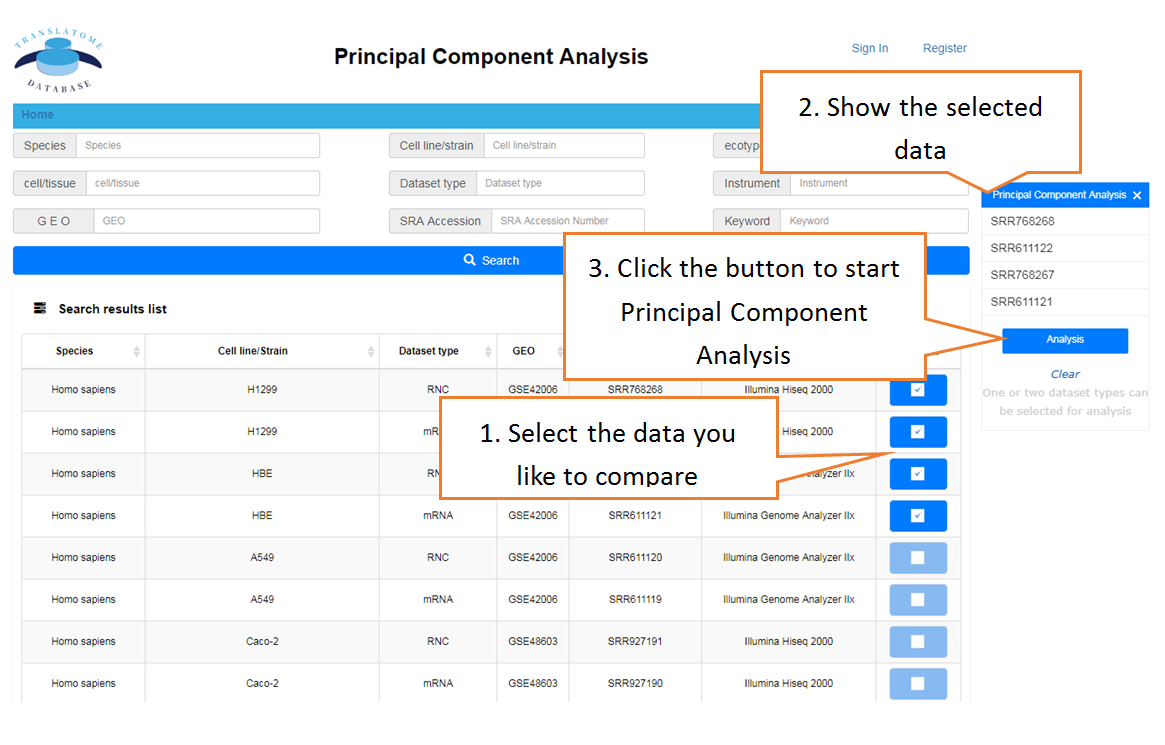
Visualize the similarity of multiple samples
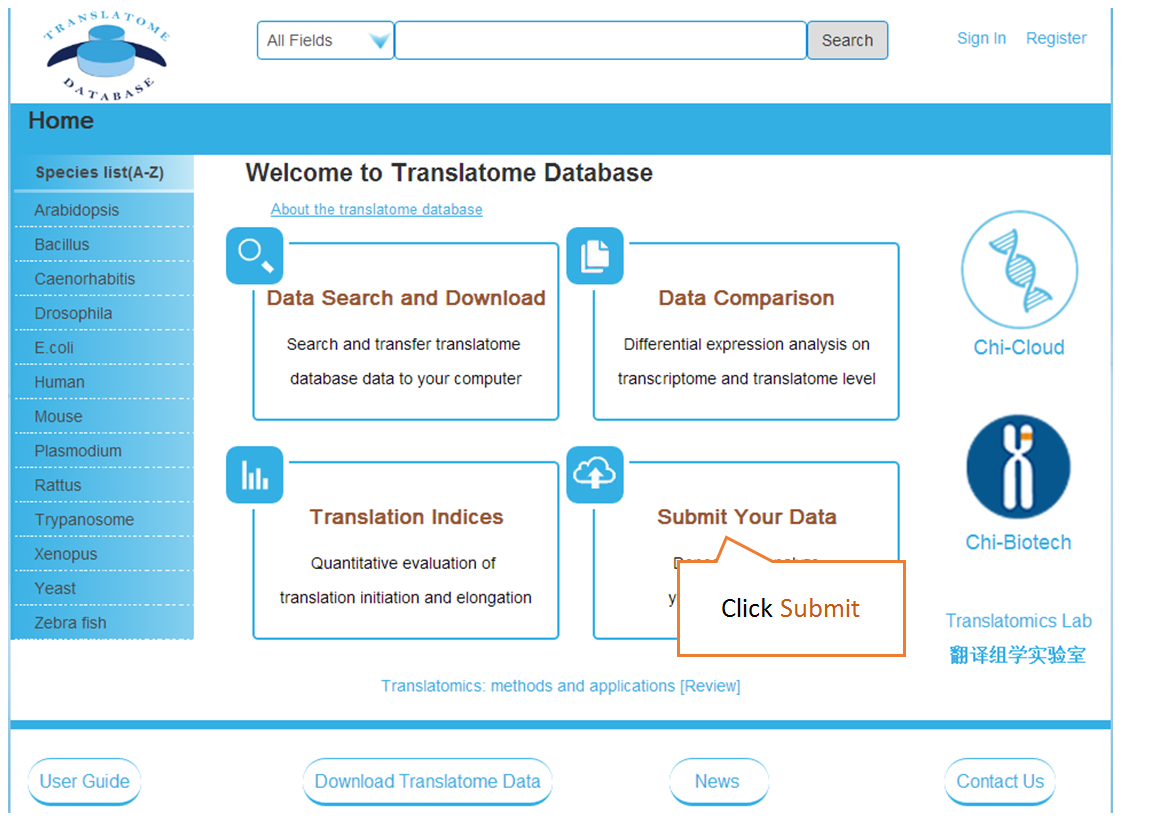
Submit Your Data: Users can submit their own translatome sequencing datasets (including RNC-seq, Ribo-seq and corresponding mRNA-seq) for analysis.
The details of transcriptome reference sequences
| Species | Version | |
|---|---|---|
| Arabidopsis | NCBI | TAIR10 RefSeq-RNA |
| Bacillus | NCBI | str.168 RefSeq-RNA |
| Caenorhabditis | UCSC | WS220/ce10 RefSeq-RNA |
| Caulobacter | NCBI | Caulobacter vibrioides CB15, RefSeq made from Genome and Gff annotation |
| Drosophila | UCSC | Dm6 RefSeq-RNA |
| E.coli | NCBI | BW25113 RefSeq-RNA |
| Human | UCSC | hg19 RefSeq-RNA |
| Leishmania | NCBI | BPK282A1 RefSeq |
| Macaca mulatta | NCBI | Mmul_10 RefSeq |
| Mouse | UCSC | mm10 RefSeq-RNA |
| Pan troglodytes | NCBI | Clint_PTRv2 RefSeq |
| Plasmodium | NCBI | 3D7 RefSeq-RNA |
| Rattus | UCSC | Rn6 RefSeq-RNA |
| Salmonella | NCBI | Enterica serovar Typhimurium str. 14028S, RefSeq made from Gff annotation and Genome |
| Solanum | NCBI | SL3.0 RefSeq |
| Soybean | NCBI | Glycine_max_v2.1 RefSeq |
| Staphylococcus | NCBI | NCTC 8325, made from Genome and Gff annotation |
| Streptomyces | NCBI | strain F613-1, , RefSeq made from Gff annotation and Genome |
| Toxoplasma | NCBI | ME49 (assembly TGA4) |
| Trypanosome | NCBI | DAL972 RefSeq-RNA |
| Vibrio Natriegens | NCBI | CCUG 16371, RefSeq made from Genome and Gff annotation |
| Xenopus | NCBI | Xenopus_laevis_v2 RefSeq-RNA |
| Yeast | NCBI | S288C RefSeq-RNA |
| Zea mays | NCBI | B73 RefGen_v4 |
| Zebrafish | UCSC | Zv9/danRer7 RefSeq-RNA |
* Download source: UCSC = UCSC Genome Browser; NCBI = NCBI Nucleotide database.
